what is the nucleotide sequence for the complementary trna
"Deoxyribonucleic acid sequencing is a outgrowth of determining Oregon identifying the order of nucleotides submit in a DNA sequence."
Nitrogenous bases, sugar and phosphate are iii ingredients of the DNA in which Adenine, T, Cytosine and Guanine are bases. A functional assemble of DNA is proverbial as a gene that encodes proteins.
For understanding the construction and function of a gene, It is very important to study its nucleotide sequence.Desoxyribonucleic acid sequencing serves this purpose. In a serial manner, the long chain of DNA is show by the sequencer automobile.
Read more:
- Structure and function of DNA
- What Is a Gene?- Definition, Structure And Function
The present article is huge, in-depth, giant and contains all the information happening DNA sequencing, how to do sequencing and different methods of DNA sequencing. Please read the article money box the end. The content of the article is,
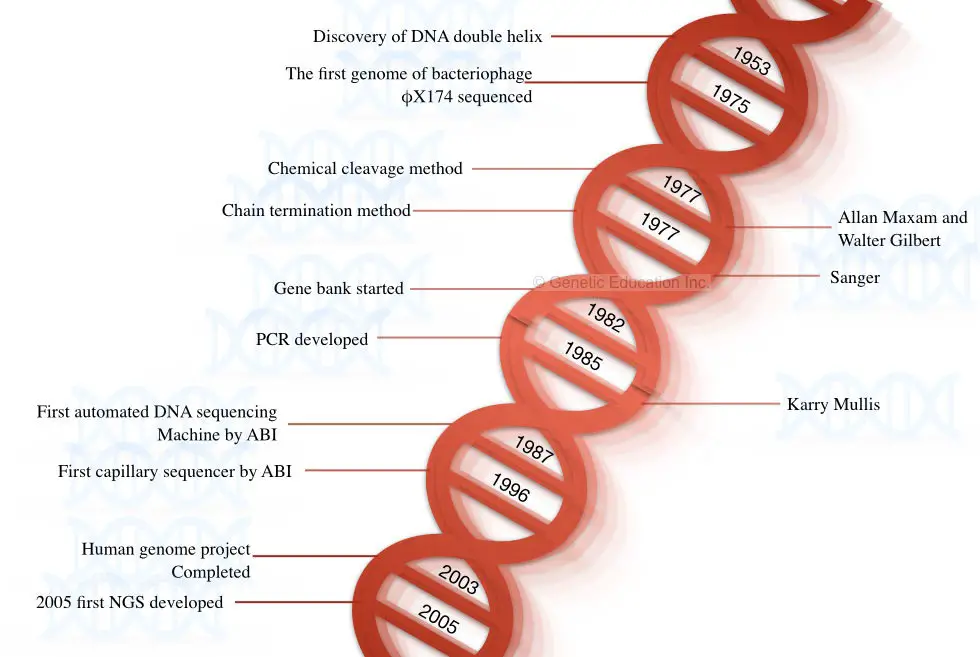
History of Deoxyribonucleic acid sequencing:
The story of DNA begins when Watson and Crick revealed the structure of DNA in the twelvemonth 1953. In 1964, Richard Holley who performed the sequencing of the tRNA was the first attempt to sequence the nucleic acid.
Using the technique of Holley and Walter Fieser, they sequenced the genome of phage MS2 (RNA sequencing). The sequenced molecules were RNA, Eventually DNA sequencing was not performed.
In the class 1977, Fredrick Sanger postulated the opening method for sequencing the DNA, named a chain termination method acting .
In the same yr, the chemical method of Desoxyribonucleic acid sequencing was explained by Allan Maxam and Walter William S. Gilbert . The genome of bacteriophage X174 was sequenced in the same year exploitation the chemical abjection method.
Because of the miss of mechanization, Both the methods (chemical degradation and chain termination) were long-winded and time-consuming. The number 1 semi-automated DNA method was developed by Lorey and Smith in the year 1986.
Afte, in the year 1987, the Practical Biosystem had developed a fully automated simple machine-controlled DNA sequencing method acting. Later the development of fully automated machines, the era of the 2000s become a golden period for sequencing platforms.
Moreover, in 1996, Practical Biosystem developed another innovative sequencing platform called body covering DNA sequencing.After that, the human genome project was completed by exploitation the combination of these methods in the year 2003.
A quick, right, reliable, and highly economic next-generation sequencing platform was postulated in the year 2005 by Solexa/Illumina. Some of the milestone into the DNA sequencing is shown in the frame below,
What is Desoxyribonucleic acid sequencing?
Studying the allelomorphic variation is not sufficient in some cases, further, using the polymerase chain reaction-equivalent methods, new polymorphism can't beryllium identified. Overcome the present problem the DNA sequencing method was evolved.
Definition:
"A testing ground technique employed to known the correct DNA chronological succession by the sequential chemical reaction is called Desoxyribonucleic acid sequencing."
laboratory processing and computational analysis some are key processes in DNA sequencing. Once the chemical substance reaction is completed, the machine-generated data are transmitted to the information processing system lab.
In the process, the amplification through by to each one nucleotide is canned in the form of signals which are collected by the machine and analyzed happening the computer. This is the basic mechanism, the nucleotides are either radio or colorful-labeled.
Now let us understand the process stepwise.
Read this article prototypic: Preparing "DNA for Sequencing".
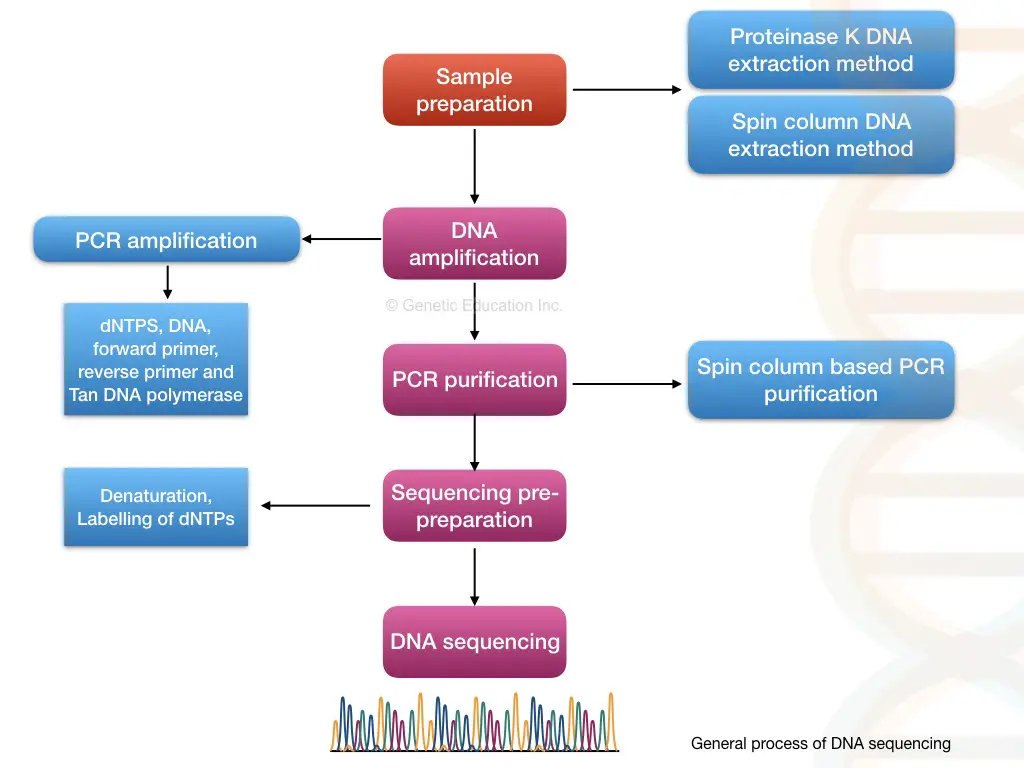
What are the stairs in DNA sequencing?
The steps mentioned to a lower place are the generalized representation of DNA sequencing, it May vary from platform to platform.
- Sample preparation (Deoxyribonucleic acid extraction)
- PCR amplification of target chronological succession
- Amplicons purification
- Sequencing pre-prep
- Deoxyribonucleic acid Sequencing
- Data analysis
Sample prep:
Present the DNA is our starting material, therefore we need to isolated DNA first. Animal, plant, microorganism, plasmid, or situation DNA can glucinium used for it.
As the prime of Deoxyribonucleic acid is the John Major concern in sequencing, we recommend using peptidase K or spin-column DNA extraction methods which have a high yield.
With the quantity, good quality of DNA also reduces the chance of response failure. Conclusively, a DNA try out with a 260/280 ratio of nearly ~1.80 (or 1.7 to 1.88) is thoughtful pristine DNA. The measure of Deoxyribonucleic acid should be ~100ng for the present assay.
PCR amplification of target sequence:
Amplification is the pivotal interfere sequencing. here only the factor of interest or the DNA sequence we wish to consider is first amplified, the rest of the DNA is discarded. Nonetheless, in whole-genome sequencing, the entire genome of an organism pot be sequenced.
A set of flanking primers temper at the outer regions of the DNA sequence of interest, therefore, unwanted DNA can't amplify. Victimization the standard PCR conditions, the amplification process is done; 35 cycles, denaturation at 94°C, tempering at 55°C to 65°C and extension at 72°C.
After the elaboration, the PCR product is run on 2% agarose colloidal gel along with the DNA ladder. Once the amplification is achieved, DNA purification is performed.
Read more:
- A Complete Guidebook of the Polymerase Chain Reaction
Amplicon purification:
Why refinement is needed?
This information is very interesting, no united will tell you virtually information technology.
If you stoppage the purity of the PCR merchandise at 260 and 280nm wavelength, information technology wish always be 1.80, exactly! because the amplicon is the pure Desoxyribonucleic acid fragment. If any contaminants are present in the DNA, elaboration will not happen. Past why is amplicon purification required? the answer is here,
Unbound primers, fuze-dimers, clean Taq DNA polymerase, clean Desoxyribonucleic acid templates, and some other new PCR reaction buffer components can abort DNA sequencing. That is why amplicon purification is required.
Here, the alcohol purification rump't work, we have to purify our PCR product with the tailspin column PCR amplicon purification kit up to reach maximum purification. After removing debris, the PCR amplicons send to the sequencing lab.
Sequencing pre-cooking:
Sampling pre-preparation is a very crucial step during DNA sequencing. During this step, adaptor Deoxyribonucleic acid sequences are ligated on both DNA ends. To the adaptor, the fuzee anneals for doing amplification. The amplification process in sequencing is connatural to PCR, however, here the nucleotides are either receiving set or colorful-labeled.
With it, high fidelity DNA polymerase and other paint ingredients are added to the reaction.
DNA sequencing:
The prepared reaction tubes are placed into the sequencer machine. During, the reaction in the sequencer, denaturation, annealing, and extension occurs, simultaneously. Here, as we are using the labeled nucleotides, the signals produced during the reaction are recorded.
The signals of the summation of each complementary nucleotide are recorded by the political machine and the data is dispatched to the computer.
Data analysis:
Once the whole DNA is sequenced, the result is redeemed into one specific file format. The intrinsic software (provided aside the manufacturing business) processed the data and compared it with the available data.
The sequence data is compared with other available data by the software program to find out variations and different mutations immediate in a factor. A abbreviated overview of the process is explained in the figure infra,
Different methods of DNA sequencing:
Various methods of DNA sequencing are explained present.
- Maxam and Gilbert method
- Ernst Boris Chain termination method acting
- semiautomated method acting
- automated method
- Pyrosequencing
- The whole-genome shotgun sequencing method
- Clone by the clone sequencing method acting
- Next-generation sequencing method
Terminologies utilised in the article:
| Term | Explanation |
| Sherd subroutine library | A collection of the entire string of the DNA (to be sequenced) fragments. |
| Gaps | Here the un-sequenced region of the DNA is called a gap. |
| Conting | A constant sequence of the Desoxyribonucleic acid assembled. |
| Say | The yield data came from the sequencer machine to the computer for one fastidious sequence. |
| Coverage | The number of multiplication the sequencing machine covered the Desoxyribonucleic acid sequence. |
Maxam-Gilbert sequencing:
The Maxam and Gilbert method was formed in 1977. It is also referred to as a chemical cleavage method acting. By using this method, they had sequenced 24 nucleotides solitary. Even so, their method was published subsequently 2 years of sangers method.
The brief principle of the present method is as stated,
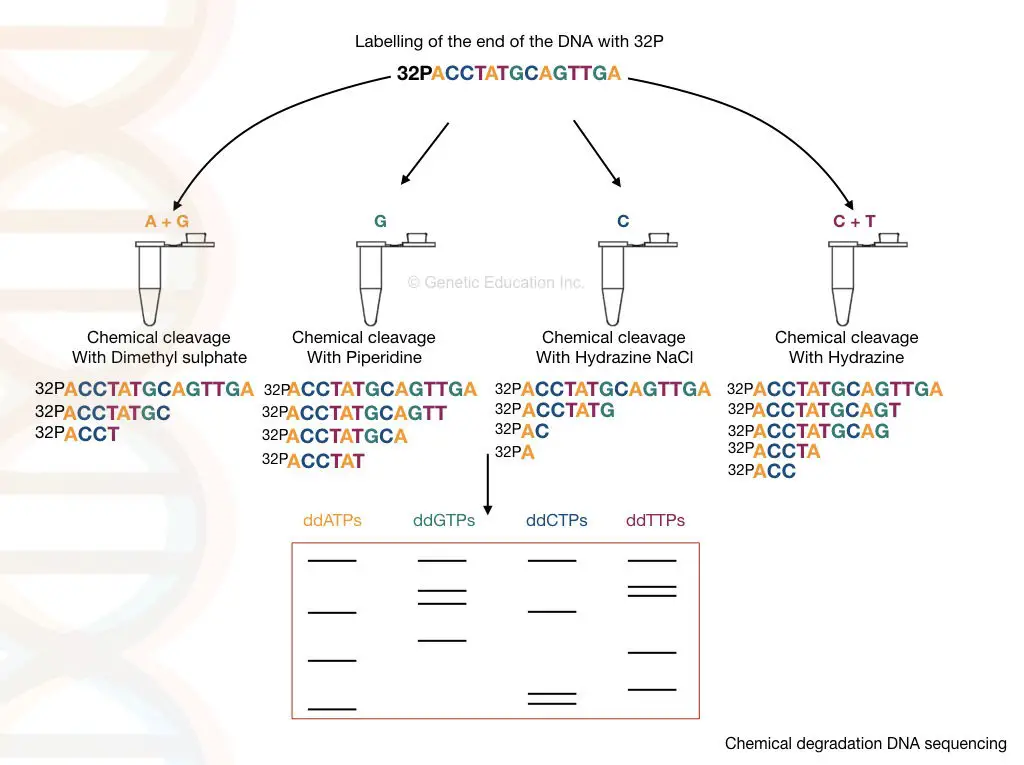
"The unshared-marooned DNA is cleaved at the specific location with the help of the chemicals at specific send location and the fragments of DNA is then incline on polyacrylamide gel."
Apparently, DNA extraction is the very first step. After that, the DNA is denatured using the rut denaturation method and single-stranded Desoxyribonucleic acid is generated.
The phosphate (5' P) conclusion of the DNA is removed and labeled by the radiolabeled P32. The enzyme onymous phosphatase removes the phosphate from the DNA and simultaneously, the kinase adds the 32P to the 5' remainder of it.
4 varied chemicals are utilized to cleave DNA at four different positions; hydrazine and hydrazine NaCl are selectively attack pyrimidine nucleotides while dimethyl sulfate and piperidine attack purine nucleotides.
- Hydrazine: T + C
- Hydrazine NaCl: C
- Dimethyl sulfate: A + G
- Piperidine: G
An equal volume of 4 different ssDNA samples is taken into 4 different tubes each containing 4 different chemicals. The samples are incubated for some prison term and electrophoresed in polyacrylamide gel electrophoresis. The results of the chemicals cleavage of four different tubes are shown in the calculate on a lower floor.
Autobiography is used to visualize the separation of DNA fragments. Due to the radiolabelled 32P final stage of the DNA, the DNA bands are visualized through autoradiography.
At our glance:
The method is more accurate than Sanger sequencing. Information technology's best suitable for DNA footprinting and DNA structural studies. IT is more advantageous over the Sanger method because the purified Deoxyribonucleic acid is directly used for sequencing.
In the represent automation time, the present method is used in DNA fingerprinting and genetic engineering studies.
Notwithstanding, plenty of disadvantages makes it harder to use regularly. The scalability of IT is poor, solitary 400bp can Be sequenced. Moreover, It is less popular because of the use of harmful radiolabeled chemicals.
Fred Sanger sequencing:
Sanger and co-workers developed a chain termination method of Deoxyribonucleic acid sequencing, after a few years of the Maxam and Gilberts method. The method is also titled the first-generation DNA sequencing method.
The chain termination method is also referred to as dideoxynucleotide sequencing because of the use of the special types of ddNTPs. The ddNTPs are antithetic from normal dNTPs. it possesses the hydrogen group instead of a hydroxyl mathematical group in the dNTPs.
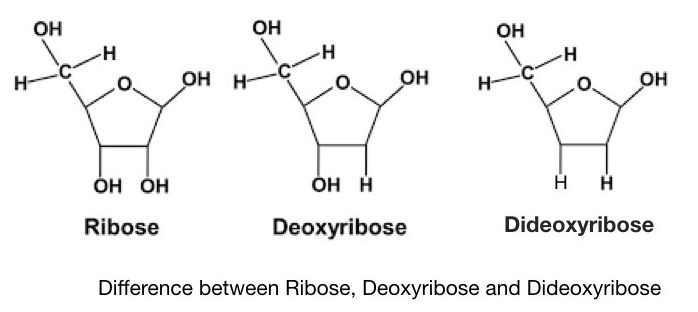
Phosphodiester bond can't form between two adjacent nucleotides referable the petit mal epilepsy of hydroxyl in ddNTPs. The nucleotide concatenation can't synthesis further and hence it is titled the chain endpoint method.
The process of Margaret Sanger sequencing is broadly divided into 3 steps:
- Deoxyribonucleic acid extraction: exploitation any of the DNA extraction protocols
- PCR amplification: exploitation the flanking primers, dNTPs, ddNTPs, Taq DNA polymerase and PCR cowcatcher.
- Identification of the amplified fragments: using autoradiography, PAGE, or capillary mousse cataphoresis.
Read more along DNA origin methods:Different types of DNA origin methods
The process of chain termination is started with DNA extraction and purification. DNA extraction can be achieved using the proteinase K method acting or the phenol-trichloromethane DNA origin method. OR we can use the ready-to-use silicon oxide-column-based kit method too.
The main aim of any DNA extraction method is to reach purity nearby ~1.8 and a amount all over 100ng.
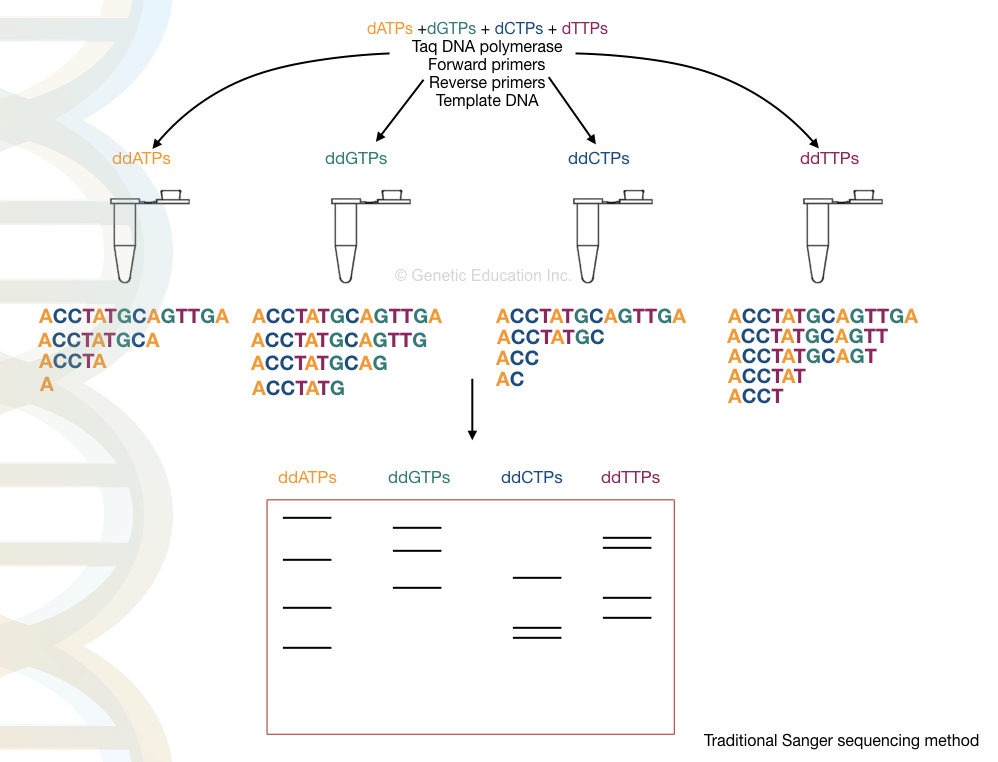
In the next step, the PCR amplification is performed past designing four distinct reactions
| Response | PCR reaction | modification |
| Reaction "A" | Taq DNA polymerase, dATPs, dGTPs, dCTPs, dGTPs and PCR buffer, primers | Tagged ddATPs |
| Response "G" | Taq Desoxyribonucleic acid polymerase, dATPs, dGTPs, dCTPs, dGTPs and PCR buffer, primers | Labeled ddGTPs |
| Response "T" | Taq Desoxyribonucleic acid polymerase, dATPs, dGTPs, dCTPs, dGTPs and PCR buffer, primers | Labeled ddTTPs |
| Chemical reaction "C" | Taq DNA polymerase, dATPs, dGTPs, dCTPs, dGTPs and PCR buffer, primers | Labeled ddCTPs |
From each one tube contains the same amount of PCR reagents only in each tube, extra ddNTPs are added as shown in the table.
The flanking primers are so much a primer that binds at the region near to our chronological sequence of involvement or the sequence that we are going to read.
Now in the next step, the Taq DNA polymerase adds the dNTPs to the DNA strand.
Hera, high faithfulness DNA polymerase is non needed.normal Taq polymerase expands the growing DNA strand by the add-on of the dNTPs. Interestingly, once it adds the ddNTP instead of dNTP the chain expansion is stopped or terminated.
The termination process is complete in 4 different tubes for 4 disparate ddNTPs. For example, in the ddATP tube, terminates the chain at all the positions where the dATPs are going to bind.
The amplified PCR products are loaded onto the polyacrylamide gel electrophoresis. The DNA fragments migrate into the colloidal gel based on the size of the fragments. The smaller fragments run for faster towards the positive charge than the larger fragments.
Before the PAGE run, the amplified DNA fragments are further denatured by heating system. Depending upon the types of labeling the gel is then analyzed under UV light or X-ray film. The banding pattern of the amplified product is shown in the calculate below,
Advancement:
The Sanger sequencing is the gold basic method acting for research as well as in the diagnosing, now. Because of its easy setup and high reproducibility. its automation had done easily.
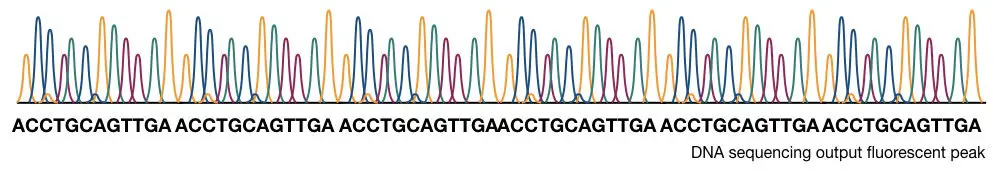
Traditionally, the results are interpreted on PAGE manually but now, the scenario is changed. The process is fully or part automated. A detector detects the fluorescence signals each time when the chain is terminated. Further, the signals are recorded and analyzed computationally.
The computational software generates various fluorescence peaks depending upon the amount of fluorescence emitted. The hypothetical illustration of the Sanger sequencing results are shown in the figure below,
How to interpret the Sri Frederick Handley Page result for Fred Sanger sequencing?
This section is very outstanding, I will teach you how you can interpret the results of the sequencing. Although it is a type of overview.
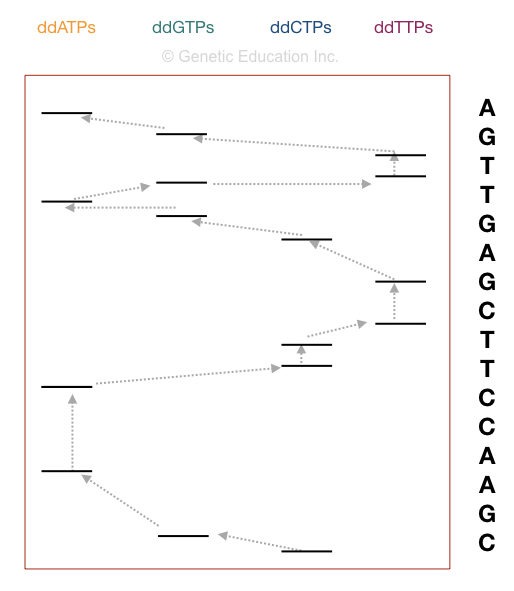
Now take a consider the figure above,
The principle of electrophoresis stated that the smaller DNA fragments migrate quicker than larger ones. Indeed the fragment on the positive side is the smallest one. Starting reading the sequence from that.
The first taper in sequencing the DNA is to match the sizing of DNA. if the bands obtained in gel and the nucleotide chronological sequence of our DNA are similar, the reaction is realised properly. For example, if your sequence length is 16, then 16 bands must comprise demo in the gel.
Get-go reading from the bottom. The Desoxyribonucleic acid sequence starts with "C" as the last fragment of Desoxyribonucleic acid expired with ddCTP. Arrange the sequencing shown in the figure, accordingly.
Machine-driven DNA sequencing:
The manual Sanger method acting was prolix. However, recent onward motio into the sequencing makes information technology easy and rapid to use. The semi-automated Sanger sequencing method acting is supported the principle of Fred Sanger's method with some minor variations.
Instead of the 4 different reactions, the automated DNA sequencing is carried out in a azygous tube. This means on a gel the Deoxyribonucleic acid runs in a single lane.
Hera in the articulated lorr-automated DNA sequencing, the fluorescent-labeled set of primers are old, alternatively of ddNTPs. Thus four different primers give quartet diametrical peaks.
The PAGE method isn't capable of separating all the fragments in a single reaction. Therefore, alternatively, the body covering gel electrophoresis method is practiced. This method separates each and every divorced fragment precisely
By using the tagged primers Beaver State the dNTPs, the machine reads the sequence accurately, on a single lane capillary electrophoresis. We dismiss sequence more than 300 samples in a single keep going an machine-controlled Desoxyribonucleic acid sequencing platform.
The capillary tubing dielectrolysis is used to separate DNA molecules on the basis of size, Information technology is powerful enough to separate a single base pair fragment. The chromatogram generated done the C.E sent the output as a fluorescent peak. Roughly, the process of information technology is explained below here,
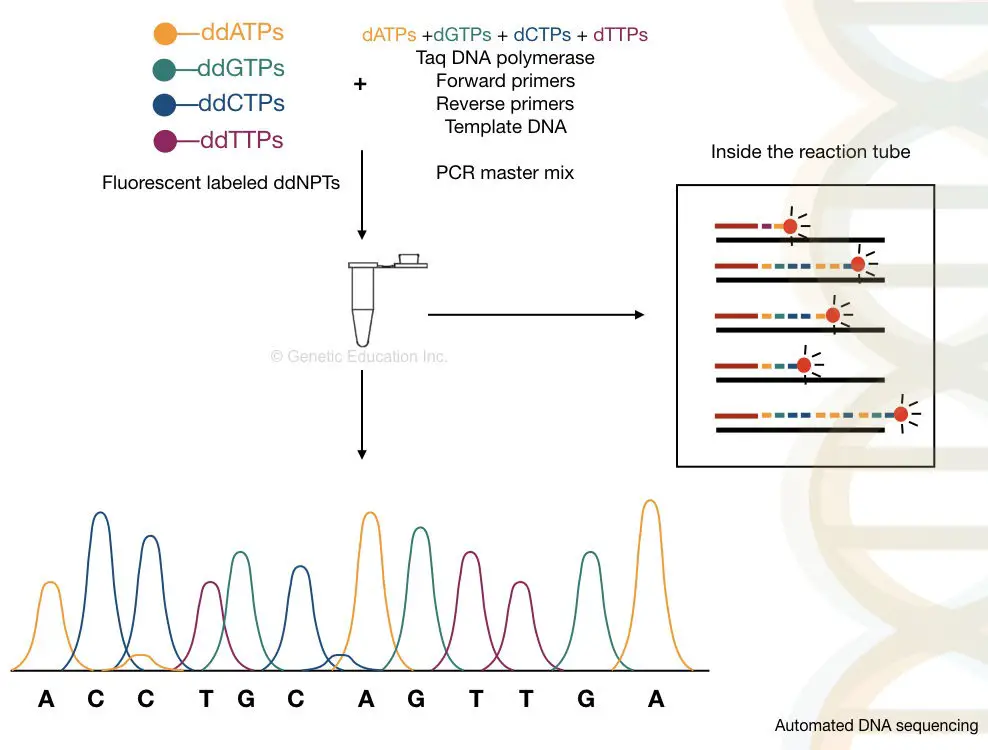
The advanced rig-automated Sanger sequencing method is much faithful, reliable and quicker than the traditional method.
What happened inside the tube?
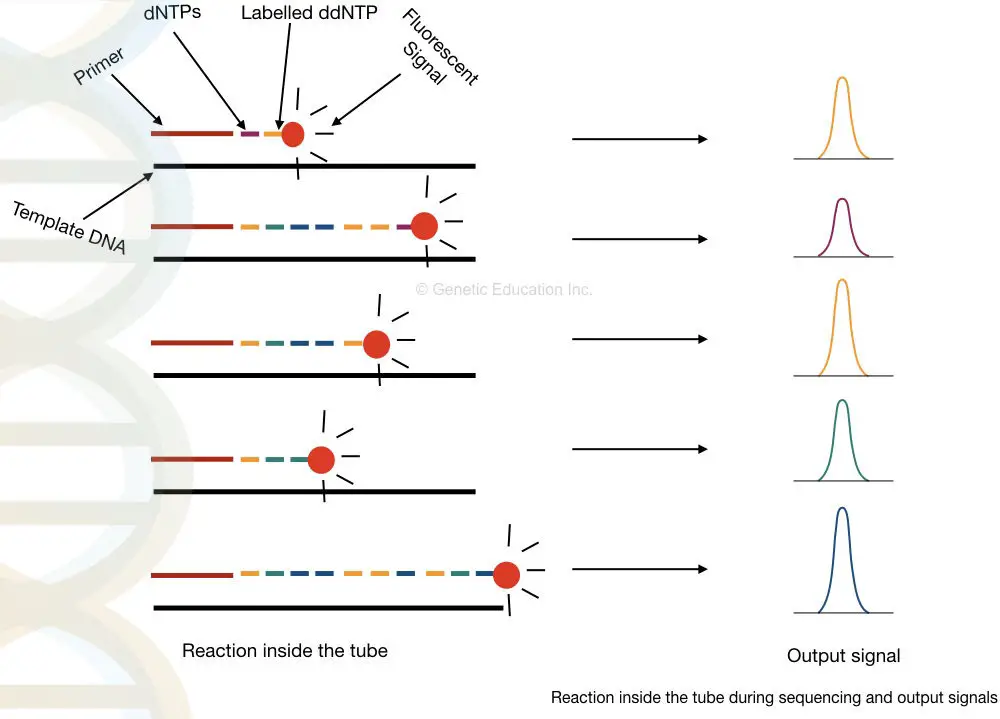
At our Glance:
The read electrical capacity of the Sanger sequencing is higher as compared with the chemical abasement method. It can sequence 700 to 800bp episode in a single run, consequently, it is Thomas More fit for sequencing bacterial or other prokaryotic genomes.
It is more advanced and automated. Smooth the error value is selfsame low as compared with the conventional chain termination method. Still, IT is time-consuming and a mellow-toll method.
Sequencing fact
It takes some 50 years to sequence all the 3.2 zillion bases of the human genome done the semi-machine-controlled Margaret Sanger sequencing method.
Pyrosequencing:
In 1993, Bertil Pettersson, Mathias Uhlen and Pål Nyren described the pyrosequencing method acting.
The method is based on the detection of the pyrophosphate released during the range reaction of nucleotide addition. Here the order of the nucleotide is determined by the PPi released during the connexion of two adjacent nucleotides (3'OH- 5'P).
In contrast with other methods, as an alternative of a single polymerase, two additional enzymes are necessary in the pyrosequencing method acting. The three enzymes are:
- DNA polymerase (without exonuclease activity)
- Luciferase
- Sulfurylase
Each three enzymes work in a sequential manner for the detection of the PPi. The sincere-time polymerase activity monitoring allows the catching of the released pyrophosphate in a cascade of the enzymatic reaction,
(DNA)n + dNTP ———————— (DNA)n+1 + PPi (Polymerase)
The addition of one dNTP removes unmatched pyrophosphate from the DNA.
PPi + APS —————————– ATP + Sol 4 -2 (ATP sulfurylase)
Adenosine triphosphate + luciferin + O2 ——————— AMP + PPi + oxyluciferin + CO2 + photon (luciferase)
Here the response is realized into the three stairs:
The enzyme polymerase adds the dNTPs to the fibre Deoxyribonucleic acid. If the correct complemental base is added, the pyrophosphate is released.
The enzyme sulfurylase converts the PPi into ATP (energy) with the help of the APS (adenosine 5´ phosphosulfate).
The ATP roleplay as a substratum for the luciferase bodily process (to a greater extent specifically "firefly luciferase"). With the help of the ATP substrate, the luciferase converts the luciferin into oxyluciferin in the presence of O and the photon of light is free.
Formerly the correct nucleotide is added, the amount of the light released by the accelerator chemical reaction is heard by the charged device coupled camera, photodiode, operating theatre a photomultiplier thermionic vacuum tube. This is the basic fundamental of the pyrosequencing frame-up.
Supported the substrate used in the proficiency ii types of pyrosequencing methods are usable: solid-phase pyroseq and liquid phase pyroseq. we leave discuss each type of pyrosequencing in both other articles.
At our Glance:
The pyrosequencing method is more accurate than the Sanger sequencing which has the mental ability to make sense to 500 nucleotides. The major vantage of pyrosequencing is the upper of the response.
However, the method required more chemical steps than a chain termination method which makes it more complex. Moreover, the read length is excessively short Eastern Samoa compared with the machine-controlled sequencing.
Whole-genome shotgun sequencing:
Eventually another modification of Sanger's chain termination method acting is the wholly-genome scattergun sequencing. However, instead of a single gene or few base pairs, the present method is powerful enough to successiveness the whole genome of an organism.
The principle of the shotgun is the assonant as Sanger's method, one additional step of DNA atomisation allows to read multiple fragments.
The full genome of an organism is divided with the help of endonuclease enzymes or by mechanical techniques. After that, the smaller fragments of DNA are sequenced individually into the machine.
The computer-supported software analyses each and every overlapping shard and reassembled it to generate the complete sequence of the entire genome.
The method can be segregated into four steps:
- Fragmentation of a DNA: with the help of restriction endonucleases or physical method acting
- Formation of libraries of the subfragments: the fragments are ligated in vectors and an entire subroutine library for various vectors are generated
- Sequencing the subfragments: from each one library is sequenced individually.
- Generation and reading the contigs: the overlapping fragments called contigs are translate aside the estimator.
The fragments generated by the lysis or restriction digestion are around 2 to 20kb.
Importantly, the shotgun sequencing reads both the sequences (it succession the double-stranded DNA) based on it contigs data, it identifies the gaps that remained unsequenced. The concise overview of present method is given to a lower place,
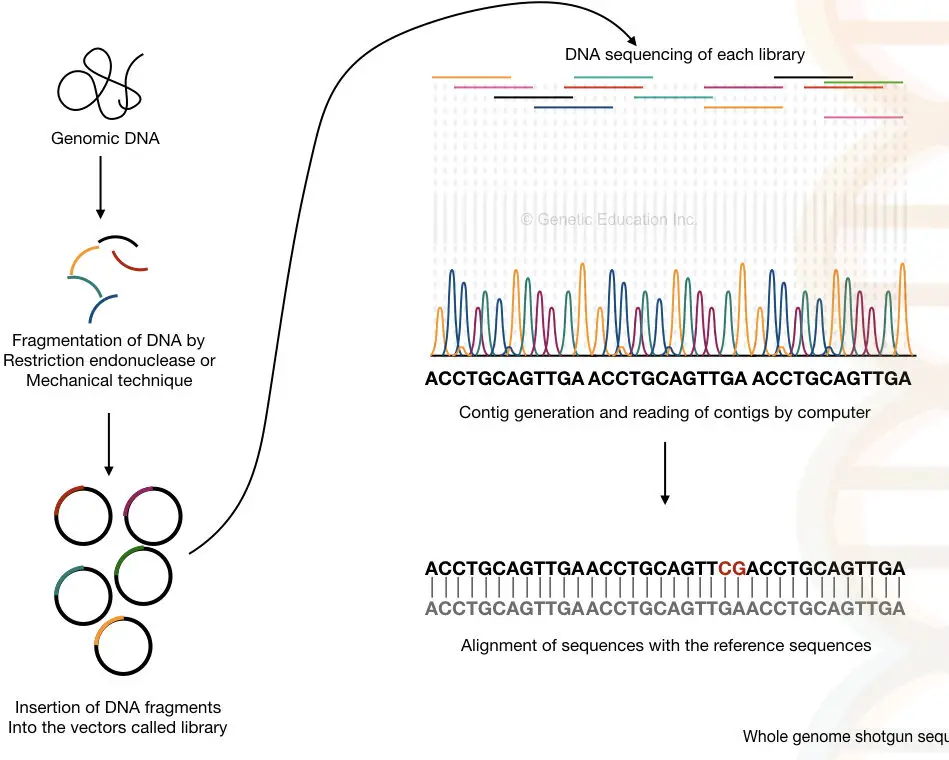
Do you know?
The scattergun sequencing concept was originally unconcealed by Sanger F and his colleagues for sequencing the whole genome.
At our glance:
The naturally occurring method is faster and cheaper than the previous technique.
The technique becomes more aggressive if any reference sequence is lendable to ordinate (it put up find out the gaps and mutations as fast as possible).
Factor or body mapping and tedious enzymatic reaction stairs are non required in the shotgun sequencing method.
Information technology has the power to sequence the overall genome of an organism (many time required to sequence the complex genome such Eastern Samoa humans and animals).
The major portion of the human genome is made high of non-functional repetitive DNA thus IT is precise unmanageable for shotgun sequencing to assembled the repetitive DNA sequences. And information technology is virtually unachievable if some denotation genome is not available.
The technique alone depends happening computational analysis. A vast, powerful, supercomputer is required to sour efficiently.
The breach can't be further filled by the techniques.
The technique becomes decrepit if reference genome sequence data is not available.
"In the year 1981, The genome of cauliflower prophet virus was sequenced aside the scattergun sequencing method."
- An Creation To Genome-Astray Association Study (GWAS).
Clone away clone sequencing:
For sequencing the healthy genome, the ringer by dead ringer method is too used instead of whole genome dead reckoning-gun sequencing. notably, this present method had helped to successfully complete the human genome jut out.
The present method is roughly similar to the previous method with one additive step. first, instead of smaller fragments, bear-sized clumps of Deoxyribonucleic acid fragments are constructed. through cistron mapping, the location of each fragment is celebrated.
Using the BACs- microorganism artificial chromosome, multiple copies of each fragment are generated for accurate sequencing. In the next step, every copied fragmentize are fragmented into smaller piece and inserted into the vectors.
The sequencing is performed as the shotgun and the lapping fragments are assembled by the computer.
In the lowest step, the data of the gene or mapping generated previously employed to assemble the sequence. the sequences are arranged connected each chromosome based on their location.
At our Glance:
Whatsoever gaps in sequencing can be known well because the fragment of the DNA is arrogated from the known locations.
Scientists can work on on various chromosomes at one time and hence a large number of DNA can live sequenced concurrently.
Techniques such as map, cloning, and restriction digestion make the present method acting more than tedious and time-consuming than old ones.
because of this argue, the present method is costlier.
Cloning the body part such every bit telomeres and centromeres are difficult to achieve which makes this proficiency eventide more toughest.
"During the yr, 1980 and 1990, the genomes of the roundworm, C. elegans and the barm, S. cerevisiae was sequenced victimisation the clone by knockoff sequencing."
Eastern Samoa the clone by clone sequencing method was used during the human genome project, IT is a break u of our history. And hence, its uncovering was one of the valuable milestones in the history of genetics.
Interestingly, the shotgun sequencing method was evolved from the clone past clone sequencing method acting.
Read much:
- DNA digital data storage
- Agarose gel electrophoresis
All chopine, we discussed Hera, has some drawbacks and limitations. Immediately, at last, the question strike in mind that, is some sequencing platform available which is fast, high-fidelity, trustworthy, veracious and cost-hard-hitting? The answer is "yes".
The next-gen sequencing, the adjacent grade of rotation in sequencing technology.
Next-generation sequencing:
The next-generation sequencing platform is different from the Sanger technique operating theatre chain termination method of DNA sequencing. Broadly, IT amplifies millions of copies of a finical shard in a massively analogue fashion and the "reads" are analyzed by the computational computer programme.
The NGS process is a trifle complex, However, it give the axe be divided into 4 different steps:
- Program library preparation
- Cluster propagation
- DNA sequencing
- Information analysis
1. Library preparation:
The library preparation is the combination of ii reactions viz, atomisation and ligation. The fragmentation of cDNA or DNA fragments is cooked by limitation digestion. After that, the smaller DNA fragments are ligated with the known DNA sequence.
The known DNA sequences are called the adapters and the process is called adapter ligation. At one time the adapters are ligated, the library of the smaller DNA fragments is generated.
The unbounded DNA fragments are washed aside the washing buffer, the appendage of library preparation is named as tagmentation.
2. Cluster generation:
What is the role of adapters??
The short oligonucleotide sequences are immobilized on the solid surface which is complemental to our adapter sequences.
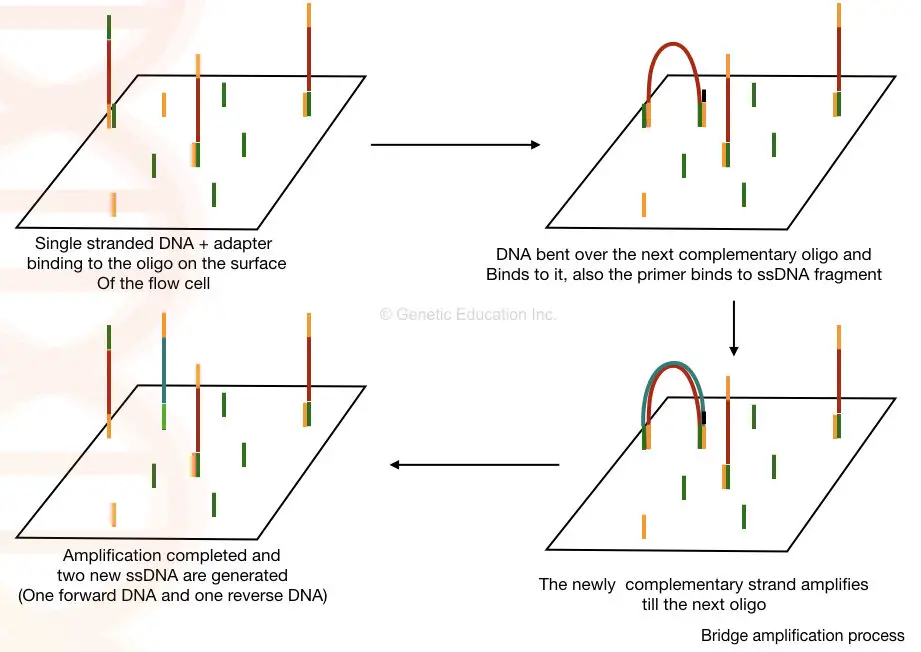
Once the library of our fragmented DNA is loaded into the cell, it is bound with the immobilized oligos connected the solid surface and by the bridge circuit amplification, the cluster of the DNA sequence is generated.
Here, in the bridge amplification, the DNA fragments bend concluded and bind to the adjacent oligo which creates a bridge. A primer binds to this Deoxyribonucleic acid sequence and is amplified vertically.
Two new unshared-stranded DNA is generated by bridge amplification. See the look-alike below,
3. Sequencing:
Eastern Samoa the polymerase adds the nucleotide into the bridge amplification, the amplification signals are recorded all time. This will generate threefold sequencing databases for the DNA sequence.
4. Data analysis:
The learn generated away the sequencing can be aligned to the reference genome chronological succession and by doing this we can identify any addition, deletion or variation into the sequence.
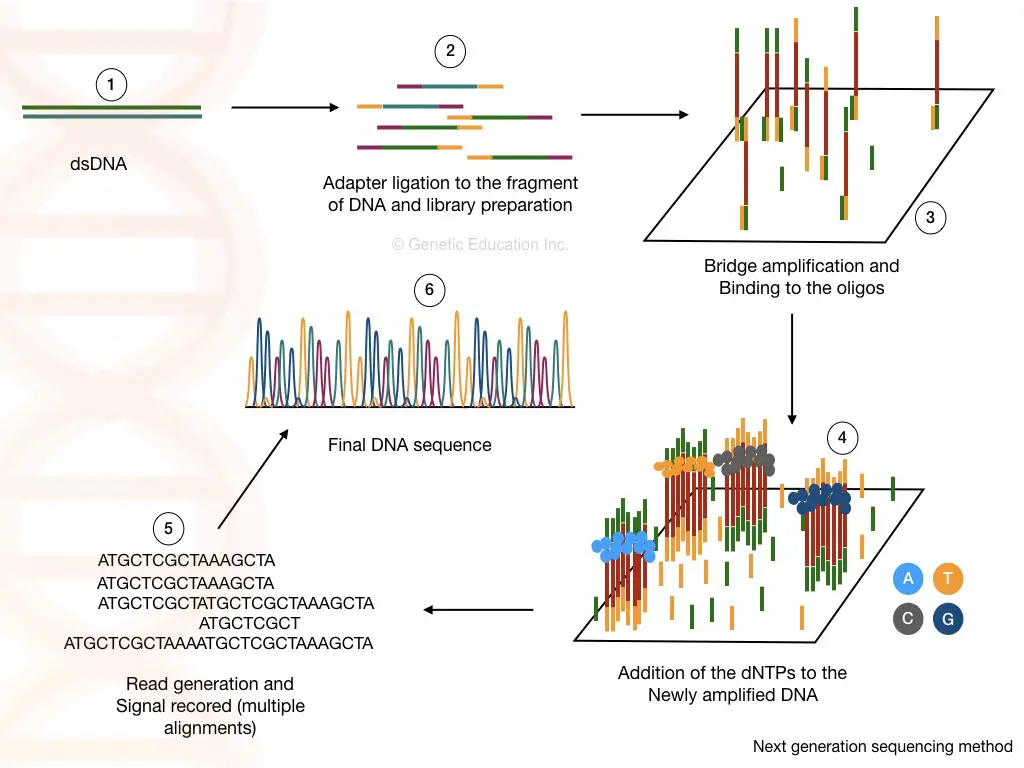
At our glance:
The NGS is the most high, fast, accurate and 100% effective technique in modern-day science.
These are the unwashed types of sequencing platforms used in genomic labs. Besides this, several other sequencing methods are:
- Single-speck real-time (RNAP) sequencing
- Individualistic-speck SMRT(Atomic number 69) sequencing
- Helioscope(Thulium) single-molecule sequencing
- DNA nano ball sequencing
- SOLiD sequencing
- Illumina (Solexa) sequencing
- Polony sequencing
- massively parallel theme song sequencing (MPSS)
- High throughput sequencing
Applications of DNA sequencing
In medical exam science, DNA sequencing potty be used in the identification of genes responsible for hereditary disorders. Current mutation can also be detected with the help of the DNA sequencing
In forensic scientific discipline, it is secondhand for maternal verification, criminal investigation and designation of individuals through with any of the available samples so much as hair, nail, blood or tissue.
In the agriculture industry, the identification of GMO species can equal possible with the helper of Deoxyribonucleic acid sequencing methods. Some minor variations in the plant genome can be detected with the help of DNA sequencing.
It is secondhand to construct maps such as whole chromosomal maps, restriction digestion maps, and genome maps. Record more happening map: A Brief Introduction to "Gene Chromosome mapping".
Open reading frames, not-open reading frames and protein-coding DNA sequences can be identified by the present method acting.
DNA sequencing is used in coding DNA/ intron, repeat chronological sequence and tandem repeat identification and detection.
Furthermore, the present method is employed in cistron manipulation and gene editing. New variations in nature can also be compulsive through sequencing.
Metagenomic studies are nowadays possible by sequencing methods such as pyrosequencing.
It is encourage used in the Microbial identification and study of the new microorganism species. The sequencing proficiency advances microbial identification by eliminating the traditional and clip-consuming culturing methods.
Nowadays microbial identification and characterization have get along more rapidly and accurately done using sequencing. Aside comparing the sequence of the target microbes with the available data, scientists can identify new mutations and new strains.
The sequencing techniques specifically, the NGS has a great application in oncology and cancer studies. various cancer-causing genes are known and characterized away the present method.
The advancement in evolutionary studies is only possible because of DNA sequencing. Aside comparing various genes and sequences, an organic process map commode be generated. Also, new variations through evolution can be encountered.
sequencing helps in perusing the asymptomatic high-risk population, prior to the natural event of disease. And thus preventive steps bathroom be taken in the first place.
Do You Know?

Sir Shankar Balasubramanian is the only known Indian scientist who was involved in the development of a sequencing platform. He was the principal investigator in the development of Solexa/ Illumina Next-generation sequencing.
Limitations of DNA sequencing
A sequencing chopine is a computer algorithm-settled assistive technique that relies connected computational data processing. For that, a huge, screechy-speed supercomputer is required.
Also, some sequences like tandem repeats, repetitive Deoxyribonucleic acid, disunited genes, and other duplicated regions can not be studied properly. The chance of errors in the pre-try out processing can cause big economical loss, these two are the major limitations of the DNA sequencing methods.
Conclusion:
John Scopes of DNA sequencing in various William Claude Dukenfield are infinite. The biggest Bob Hope of it is its use in the diagnosing of multigenic genetic disorders. The present method commode be a useful tool for antenatal and preimplantation genetic studies.
In the latest scientific trends, the development of factor therapies and the concept of personalized medicines can not be fulfilled without NGS-like robust techniques. However, more optimization and advancements are required to reduce the cots, time-length and erroneousness rate in the present genetic technology.
Source:
- Brown TA. Genomes. 2nd edition. Oxford: Wiley-Liss; 2002. Chapter 6, Sequencing Genomes.
- Heather JM, Range B. The sequence of sequencers: The story of sequencing DNA.Genomics. 2016;107(1):1-8.
- Lehrach H. DNA sequencing methods in human genetic science and disease research.F1000Prime Rep. 2013;5:34. Published 2013 September 2.
what is the nucleotide sequence for the complementary trna
Source: https://geneticeducation.co.in/dna-sequencing-history-steps-methods-applications-and-limitations/
Posting Komentar untuk "what is the nucleotide sequence for the complementary trna"